Tazobactam pdb setup
The vm1_taz.pdb file does not have hydrogens.
 |
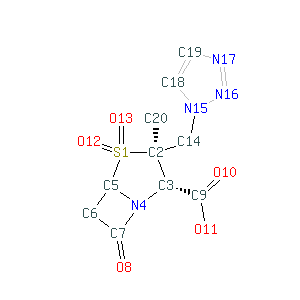 |
| 1vm1 TAZ structure |
Chemical structure |
Without the hydrogens,
it is not possible to determine the bond orders and come up with
parameters for modeling. There are several possibilities for adding
in hydrogens:
- use xleap editor to draw in hydrogens using the published
chemical structure showing bond orders.
- use a ligand index such as
"Macromolecular Structure Database" (MSD)
- edit in guesses to the H atoms and their positions
by hand in the pdb.
Using the MSD:
- Type in TAZ or taz in the three-letter-code box
or type in tazobactam in the molecule name box
and click SEARCH button
- Click TAZ on left side:
make sure:
- Output is "text"
- Format is "pdb"
- Library is "ideal"
- Hydrogens box MUST BE CHECKED!!!!!
- click Save button
- navigate your way to the dir where you want to save the file
- rename filename from cgi.pl to TAZ.pdb and Save
This structure is ready for antechamber to determine parameters necessary
for modeling the molecule in Amber. After determining the parameters
in antechamber, there is no more need for this pdb file.




